Reads all configuration tables and other model data from a location
where it was previously compiled to by the compile function.
Value
Compiled slendr_model model object which encapsulates all
information about the specified model (which populations are involved,
when and how much gene flow should occur, what is the spatial resolution
of a map, and what spatial dispersal and mating parameters should be used
in a SLiM simulation, if applicable)
Examples
check_dependencies(python = TRUE, quit = TRUE) # dependencies must be present
init_env()
#> The interface to all required Python modules has been activated.
# load an example model with an already simulated tree sequence
path <- system.file("extdata/models/introgression", package = "slendr")
model <- read_model(path)
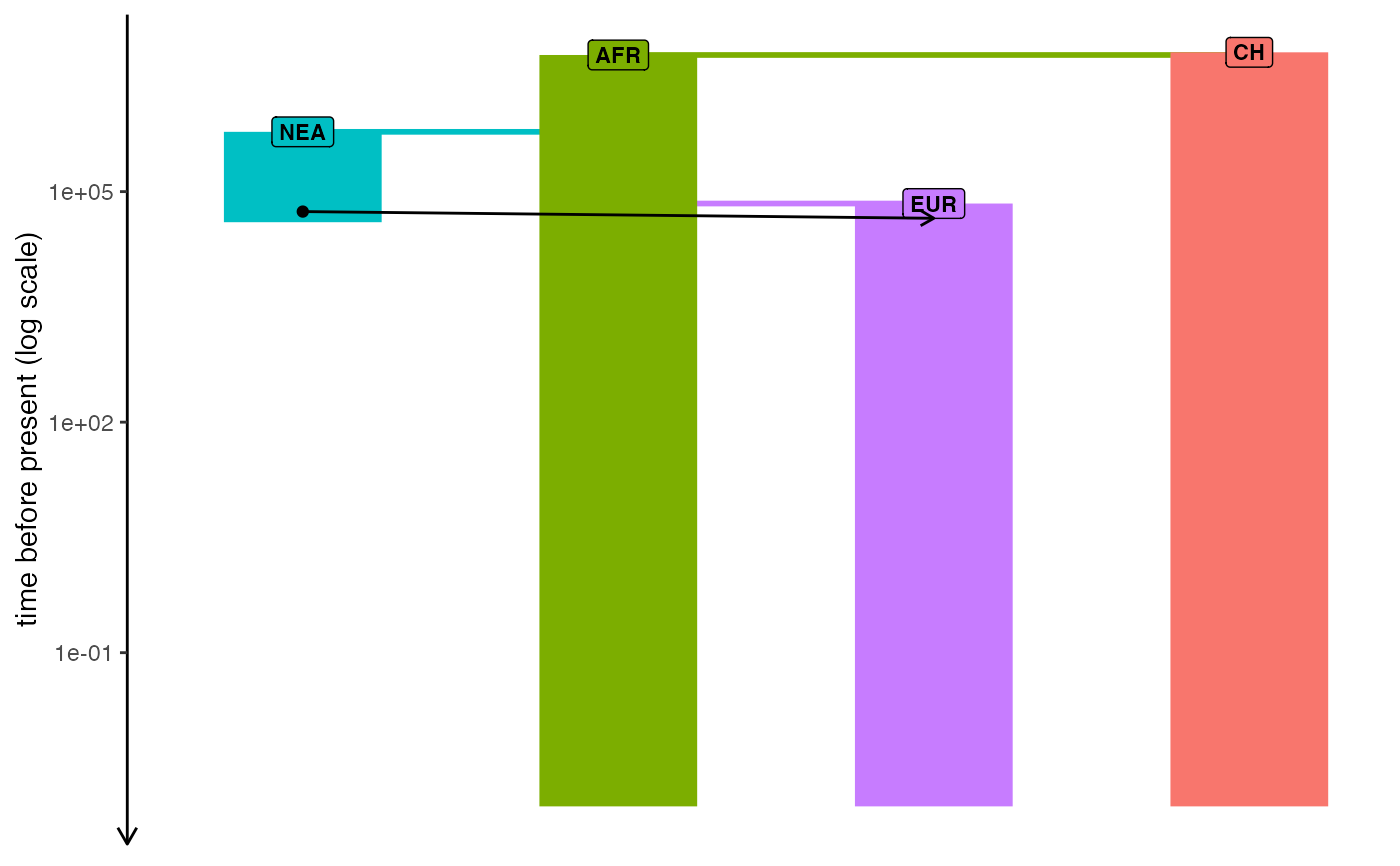
plot_model(model, sizes = FALSE, log = TRUE)